Using the ScPCAr package to access Single-cell Pediatric Cancer Atlas data
Source:vignettes/ScPCAr.Rmd
ScPCAr.RmdIntroduction
The ScPCAr package provides an interface to interact with the Single-cell Pediatric Cancer Atlas (ScPCA) Portal API. This vignette demonstrates the basic workflow for discovering, downloading, and working with ScPCA data.
This vignette covers the following tasks for interacting with the ScPCA Portal:
- Listing available projects
- Selecting a project and exploring its samples
- Obtaining an authentication token
- Downloading data for individual samples
- Loading the data into R
Installing the ScPCAr Package
The ScPCAr package is currently available via GitHub. You can install
the latest version using the remotes package:
# Install remotes if needed
if (!requireNamespace("remotes", quietly = TRUE)) {
install.packages("remotes")
}
# Install ScPCAr from GitHub
remotes::install_github("Alexslemonade/ScPCAr")You can then load the ScPCAr package. Note that in this
vignette we will use ScPCAr:: when calling functions from
the package, both for clarity and to avoid any possible namespace
conflicts. We will also load the SingleCellExperiment
package for later analysis.
Exploring available projects
Listing all projects
First, let’s see what projects are available in the ScPCA Portal:
# Get a data frame of all projects
projects <- ScPCAr::scpca_projects()
# print out a portion of the data frame
head(projects)## # A tibble: 6 × 23
## scpca_project_id sample_count title pi_name abstract additional_restricti…¹
## <chr> <int> <chr> <chr> <chr> <chr>
## 1 SCPCP000001 23 Single … green_… Pediatr… Research or academic …
## 2 SCPCP000002 26 Single … green_… Pediatr… Research or academic …
## 3 SCPCP000003 59 Single … teache… Early T… Research or academic …
## 4 SCPCP000006 45 Single … murphy… Wilms t… Research or academic …
## 5 SCPCP000007 30 Single-… gawad Bulk ge… Research or academic …
## 6 SCPCP000008 104 Single-… mullig… Acute l… Research or academic …
## # ℹ abbreviated name: ¹additional_restrictions
## # ℹ 17 more variables: created_at <dttm>, downloadable_sample_count <int>,
## # has_bulk_rna_seq <lgl>, has_cite_seq_data <lgl>,
## # has_multiplexed_data <lgl>, has_single_cell_data <lgl>,
## # has_spatial_data <lgl>, human_readable_pi_name <chr>,
## # includes_anndata <lgl>, includes_cell_lines <lgl>,
## # includes_merged_anndata <lgl>, includes_merged_sce <lgl>, …The scpca_projects() function returns a data frame with
basic project metadata. By default, it returns a simplified version with
list columns removed for easier viewing. You can see the full structure
with additional data such as a list of diagnoses, data types, external
accession numbers, etc., by setting simplify = FALSE:
# Get the full project information including list columns
projects_full <- ScPCAr::scpca_projects(simplify = FALSE)
# View the structure of the full data frame
dplyr::glimpse(projects_full)## Rows: 23
## Columns: 37
## $ scpca_project_id <chr> "SCPCP000001", "SCPCP000002", "SCPCP000003",…
## $ sample_count <int> 23, 26, 59, 45, 30, 104, 39, 42, 11, 33, 10,…
## $ title <chr> "Single cell RNA sequencing of pediatric hig…
## $ pi_name <chr> "green_mulcahy_levy", "green_mulcahy_levy", …
## $ abstract <chr> "Pediatric brain tumors are now the most com…
## $ additional_metadata_keys <list> <"development_stage_ontology_term_id", "dis…
## $ additional_restrictions <chr> "Research or academic purposes only", "Resea…
## $ computed_files <list> [<data.frame[5 x 17]>], [<data.frame[5 x 17…
## $ contacts <list> [<data.frame[1 x 2]>], [<data.frame[1 x 2]>…
## $ created_at <dttm> 2025-08-26, 2025-08-26, 2025-08-26, 2025-08…
## $ diagnoses_counts <df[,56]> <data.frame[23 x 56]>
## $ diagnoses <list> <"Anaplastic astrocytoma", "Anaplastic g…
## $ disease_timings <list> <"Metastatic recurrence of anaplastic pleom…
## $ downloadable_sample_count <int> 23, 26, 59, 43, 30, 104, 38, 42, 11, 33, 10…
## $ external_accessions <list> [<data.frame[4 x 3]>], [<data.frame[0 x 0]>]…
## $ has_bulk_rna_seq <lgl> TRUE, TRUE, TRUE, TRUE, FALSE, FALSE, TRUE,…
## $ has_cite_seq_data <lgl> FALSE, FALSE, TRUE, FALSE, TRUE, TRUE, FALSE…
## $ has_multiplexed_data <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, TR…
## $ has_single_cell_data <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TR…
## $ has_spatial_data <lgl> FALSE, FALSE, FALSE, TRUE, FALSE, FALSE, FAL…
## $ human_readable_pi_name <chr> "Green/Mulcahy Levy", "Green/Mulcahy Levy", …
## $ includes_anndata <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TR…
## $ includes_cell_lines <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FA…
## $ includes_merged_anndata <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, FALSE, FALSE, …
## $ includes_merged_sce <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, FALSE, FALSE, …
## $ includes_xenografts <lgl> FALSE, FALSE, TRUE, FALSE, FALSE, FALSE, FAL…
## $ metadata_dataset_id <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
## $ modalities <list> <"SINGLE_CELL", "BULK_RNA_SEQ">, <"SINGLE_CE…
## $ multiplexed_sample_count <int> 0, 0, 0, 0, 0, 0, 34, 0, 0, 0, 0, 0, 0, 0, …
## $ organisms <list> "Homo sapiens", "Homo sapiens", "Homo sapien…
## $ publications <list> [<data.frame[1 x 3]>], [<data.frame[0 x 0]>…
## $ samples <list> <"SCPCS000001", "SCPCS000002", "SCPCS000003…
## $ seq_units <list> "cell", "cell", "cell", <"nucleus", "spot">…
## $ summaries <list> [<data.frame[7 x 5]>], [<data.frame[5 x 5]>…
## $ technologies <list> "10Xv3", "10Xv3", "10Xv3", <"10Xv3.1", "vis…
## $ unavailable_samples_count <int> 0, 0, 0, 2, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0…
## $ updated_at <dttm> 2025-08-26, 2025-08-26, 2025-08-26, 2025-08-…Getting detailed project information
Now let’s get more detailed information about the samples in a
selected project. We will use the first project,
SCPCP000001, as an example. According to the project info
above, these samples are from a study of pediatric high-grade gliomas.
We can get more detailed information about this project using its
project_id, and here we will set
simplifyVector = TRUE to convert simple lists into vectors
where possible.
project_id <- "SCPCP000001"
# Get detailed project metadata
project_info <- ScPCAr::get_project_info(project_id)This returns a list with more detailed information about
the project and the samples within. You can explore the full structure
of this list with str() or dplyr::glimpse(),
but for now we will just look at a few of the components that might be
of interest.
For example, we can look at the set of diagnoses that are present, and their counts:
data.frame(count = unlist(project_info$diagnoses_counts)) |>
tibble::rownames_to_column("diagnosis")## diagnosis count
## 1 Glioblastoma 16
## 2 Non-cancerous 1
## 3 Anaplastic glioma 1
## 4 High-grade glioma 2
## 5 Anaplastic astrocytoma 1
## 6 Diffuse midline glioma 1
## 7 Pleomorphic xanthoastrocytoma 1Similarly, we can look at the data modalities available for this project, which here includes both single-cell and bulk RNA-seq data:
project_info$modalities## [1] "SINGLE_CELL" "BULK_RNA_SEQ"Exploring sample information
Getting sample metadata
Let’s look at the samples within our selected project:
# Get sample information for the project
samples <- ScPCAr::get_project_samples(project_id)
head(samples)## # A tibble: 6 × 34
## scpca_sample_id scpca_project_id outcome organism submitter_id participant_id
## <chr> <chr> <chr> <chr> <chr> <chr>
## 1 SCPCS000001 SCPCP000001 PFS 11 … Homo sa… 834 834
## 2 SCPCS000002 SCPCP000001 PFS 11 … Homo sa… 1107 1107
## 3 SCPCS000003 SCPCP000001 PFS 7 m… Homo sa… 1115 1115
## 4 SCPCS000004 SCPCP000001 PFS 3 m… Homo sa… 1431 1431
## 5 SCPCS000005 SCPCP000001 PFS 3 m… Homo sa… 1437 1437
## 6 SCPCS000006 SCPCP000001 Stable … Homo sa… 1458 1458
## # ℹ 28 more variables: organism_ontology_id <chr>, sex_ontology_term_id <chr>,
## # tissue_ontology_term_id <chr>, disease_ontology_term_id <chr>,
## # molecular_characteristics <chr>, spinal_leptomeningeal_mets <chr>,
## # development_stage_ontology_term_id <chr>,
## # self_reported_ethnicity_ontology_term_id <chr>, age <dbl>,
## # age_timing <chr>, created_at <dttm>, demux_cell_count_estimate_sum <lgl>,
## # diagnosis <chr>, disease_timing <chr>, has_bulk_rna_seq <lgl>, …Getting detailed sample information
We can also get detailed information for a specific sample:
sample_id <- "SCPCS000001"
# Get detailed sample metadata
sample_detail <- ScPCAr::get_sample_info(sample_id)
str(sample_detail, max.level = 1)## List of 28
## $ additional_metadata :List of 12
## $ age : chr "14"
## $ age_timing : chr "diagnosis"
## $ computed_files :'data.frame': 2 obs. of 17 variables:
## $ created_at : chr "2025-08-26T07:31:52.502575Z"
## $ demux_cell_count_estimate_sum: NULL
## $ diagnosis : chr "Anaplastic glioma"
## $ disease_timing : chr "Initial diagnosis"
## $ has_bulk_rna_seq : logi TRUE
## $ has_cite_seq_data : logi FALSE
## $ has_multiplexed_data : logi FALSE
## $ has_single_cell_data : logi TRUE
## $ has_spatial_data : logi FALSE
## $ includes_anndata : logi TRUE
## $ is_cell_line : logi FALSE
## $ is_xenograft : logi FALSE
## $ modalities : chr [1:2] "SINGLE_CELL" "BULK_RNA_SEQ"
## $ multiplexed_with : list()
## $ project :List of 37
## $ sample_cell_count_estimate : int 3422
## $ scpca_id : chr "SCPCS000001"
## $ seq_units : chr [1:2] "bulk" "cell"
## $ sex : chr "F"
## $ subdiagnosis : chr "NA"
## $ technologies : chr [1:2] "10Xv3" "paired_end"
## $ tissue_location : chr "Right thalamus/midbrain"
## $ treatment : chr "Debulking, RT, irinotecan/cetuximab"
## $ updated_at : chr "2025-08-26T07:31:52.502598Z"
# Check which data modalities are available for this sample
sample_detail$modalities## [1] "SINGLE_CELL" "BULK_RNA_SEQ"Authentication
To download actual data files, you need an agree to the terms of
service and obtain an authentication token. To view the terms of service
in a web browser, run view_terms(). Then you can use the
get_auth() function, providing your email address and
agreeing to the terms of service:
# Get an authentication token
# Replace with your actual email address
my_email <- "your.email@example.com"
auth_token <- ScPCAr::get_auth(email = my_email, agree = TRUE)Important Notes:
- You must set
agree = TRUEto indicate you accept the terms of service - Replace
"your.email@example.com"with your actual email address - Keep your token secure and don’t share it publicly
Downloading data for an ScPCA sample
Downloading SingleCellExperiment objects
Now we can download data for our selected sample. We will use the
download_sample() function, specifying the sample ID,
authentication token, desired destination directory, and the file
format. The function will download and unpack the files associated with
that sample, and return a list of file paths for the downloaded files
Let’s start with SingleCellExperiment format:
# Download SingleCellExperiment data for our sample
# This will create a directory structure under "scpca_data/"
file_paths <- ScPCAr::download_sample(
sample_id = "SCPCS000001",
auth_token = auth_token,
destination = "scpca_data",
format = "sce"
)
# List the files that were downloaded
file_paths## [1] "scpca_data/SCPCS000001_SINGLE-CELL_SINGLE-CELL-EXPERIMENT_2025-10-17/README.md"
## [2] "scpca_data/SCPCS000001_SINGLE-CELL_SINGLE-CELL-EXPERIMENT_2025-10-17/SCPCL000001_celltype-report.html"
## [3] "scpca_data/SCPCS000001_SINGLE-CELL_SINGLE-CELL-EXPERIMENT_2025-10-17/SCPCL000001_filtered.rds"
## [4] "scpca_data/SCPCS000001_SINGLE-CELL_SINGLE-CELL-EXPERIMENT_2025-10-17/SCPCL000001_processed.rds"
## [5] "scpca_data/SCPCS000001_SINGLE-CELL_SINGLE-CELL-EXPERIMENT_2025-10-17/SCPCL000001_qc.html"
## [6] "scpca_data/SCPCS000001_SINGLE-CELL_SINGLE-CELL-EXPERIMENT_2025-10-17/SCPCL000001_unfiltered.rds"
## [7] "scpca_data/SCPCS000001_SINGLE-CELL_SINGLE-CELL-EXPERIMENT_2025-10-17/single_cell_metadata.tsv"Understanding the downloaded file structure
A standard download of SingleCellExperiment data for a sample will create a directory structure like this:
scpca_data/
└── {sample_id}_{MODALITY}_{FORMAT}_{YYYY-MM-DD}/
├── README.md
├── single_cell_metadata.tsv
├── {library_id}_filtered.rds
├── {library_id}_processed.rds
├── {library_id}_unfiltered.rds
├── {library_id}_qc.html
└── {library_id}_celltype-report.htmlNote that the library_id and sample_id
often have different numbers, as a single sample may have been sequenced
across multiple libraries. For details about the contents of these
files, see the ScPCA
Portal documentation.
Loading data into R
Now let’s load in the processed data for our sample. First we will
select the processed SingleCellExperiment file from the downloaded
paths, then use readRDS() to load it.
# select the processed SCE file using a pattern match for the file name.
processed_sce_files <- stringr::str_subset(file_paths, "_processed\\.rds$")
# Load the first (in this case only) SingleCellExperiment object
sce <- readRDS(processed_sce_files[1])
# View a summary of the object
sce## class: SingleCellExperiment
## dim: 60319 2628
## metadata(40): library_id sample_id ... cellassign_reference_version
## cellassign_reference_organs
## assays(3): counts spliced logcounts
## rownames(60319): ENSG00000223972 ENSG00000243485 ... ENSG00000273496
## ENSG00000274175
## rowData names(4): gene_ids gene_symbol mean detected
## colnames(2628): GTTCGCTGTTCTCTCG CTCTCAGCATGGATCT ... TCATACTTCTGCGAGC
## TCTCACGAGGACGGAG
## colData names(19): barcodes sum ... consensus_celltype_annotation
## consensus_celltype_ontology
## reducedDimNames(2): PCA UMAP
## mainExpName: NULL
## altExpNames(0):Working with the SingleCellExperiment object
Once loaded, you can work with the SingleCellExperiment object using standard Bioconductor tools. For more information on the contents of the SingleCellExperiment objects provided by ScPCA, see the ScPCA Portal documentation.
## [1] 60319 2628## DataFrame with 6 rows and 19 columns
## barcodes sum detected subsets_mito_sum
## <character> <numeric> <integer> <numeric>
## GTTCGCTGTTCTCTCG GTTCGCTGTTCTCTCG 57012 9097 1494
## CTCTCAGCATGGATCT CTCTCAGCATGGATCT 52113 8625 4064
## TCAGCCTCAGGTATGG TCAGCCTCAGGTATGG 49759 8650 5253
## CCTTGTGGTCCATAGT CCTTGTGGTCCATAGT 58121 9177 3540
## AGCGCCATCTTCGACC AGCGCCATCTTCGACC 51639 8570 5607
## CTGTAGATCCATAGGT CTGTAGATCCATAGGT 53179 8962 6712
## subsets_mito_detected subsets_mito_percent total
## <integer> <numeric> <numeric>
## GTTCGCTGTTCTCTCG 13 2.62050 57012
## CTCTCAGCATGGATCT 14 7.79844 52113
## TCAGCCTCAGGTATGG 14 10.55688 49759
## CCTTGTGGTCCATAGT 14 6.09074 58121
## AGCGCCATCTTCGACC 14 10.85807 51639
## CTGTAGATCCATAGGT 14 12.62152 53179
## prob_compromised miQC_pass scpca_filter sizeFactor cluster
## <numeric> <logical> <character> <numeric> <factor>
## GTTCGCTGTTCTCTCG 6.09745e-06 TRUE Keep 5.03806 1
## CTCTCAGCATGGATCT 8.68181e-06 TRUE Keep 3.50519 1
## TCAGCCTCAGGTATGG 2.67246e-06 TRUE Keep 3.59865 1
## CCTTGTGGTCCATAGT 7.71935e-07 TRUE Keep 4.48946 1
## AGCGCCATCTTCGACC 3.74199e-06 TRUE Keep 3.38054 1
## CTGTAGATCCATAGGT 2.00608e-07 TRUE Keep 3.53366 1
## singler_celltype_ontology singler_celltype_annotation
## <character> <character>
## GTTCGCTGTTCTCTCG CL:0000540 neuron
## CTCTCAGCATGGATCT CL:0000540 neuron
## TCAGCCTCAGGTATGG CL:0000540 neuron
## CCTTGTGGTCCATAGT CL:0000540 neuron
## AGCGCCATCTTCGACC CL:0000540 neuron
## CTGTAGATCCATAGGT CL:0000540 neuron
## cellassign_celltype_annotation cellassign_celltype_ontology
## <character> <character>
## GTTCGCTGTTCTCTCG Gamma delta T cells CL:0000798
## CTCTCAGCATGGATCT Purkinje neurons CL:0000121
## TCAGCCTCAGGTATGG Purkinje neurons CL:0000121
## CCTTGTGGTCCATAGT Gamma delta T cells CL:0000798
## AGCGCCATCTTCGACC Purkinje neurons CL:0000121
## CTGTAGATCCATAGGT Purkinje neurons CL:0000121
## cellassign_max_prediction consensus_celltype_annotation
## <numeric> <character>
## GTTCGCTGTTCTCTCG 1.000000 Unknown
## CTCTCAGCATGGATCT 1.000000 neuron
## TCAGCCTCAGGTATGG 0.963709 neuron
## CCTTGTGGTCCATAGT 1.000000 Unknown
## AGCGCCATCTTCGACC 1.000000 neuron
## CTGTAGATCCATAGGT 1.000000 neuron
## consensus_celltype_ontology
## <character>
## GTTCGCTGTTCTCTCG NA
## CTCTCAGCATGGATCT CL:0000540
## TCAGCCTCAGGTATGG CL:0000540
## CCTTGTGGTCCATAGT NA
## AGCGCCATCTTCGACC CL:0000540
## CTGTAGATCCATAGGT CL:0000540## DataFrame with 6 rows and 4 columns
## gene_ids gene_symbol mean detected
## <character> <character> <numeric> <numeric>
## ENSG00000223972 ENSG00000223972 DDX11L1 0.000380518 0.0380518
## ENSG00000243485 ENSG00000243485 MIR1302-2HG 0.000000000 0.0000000
## ENSG00000284332 ENSG00000284332 MIR1302-2 0.000000000 0.0000000
## ENSG00000268020 ENSG00000268020 OR4G4P 0.000000000 0.0000000
## ENSG00000240361 ENSG00000240361 OR4G11P 0.000000000 0.0000000
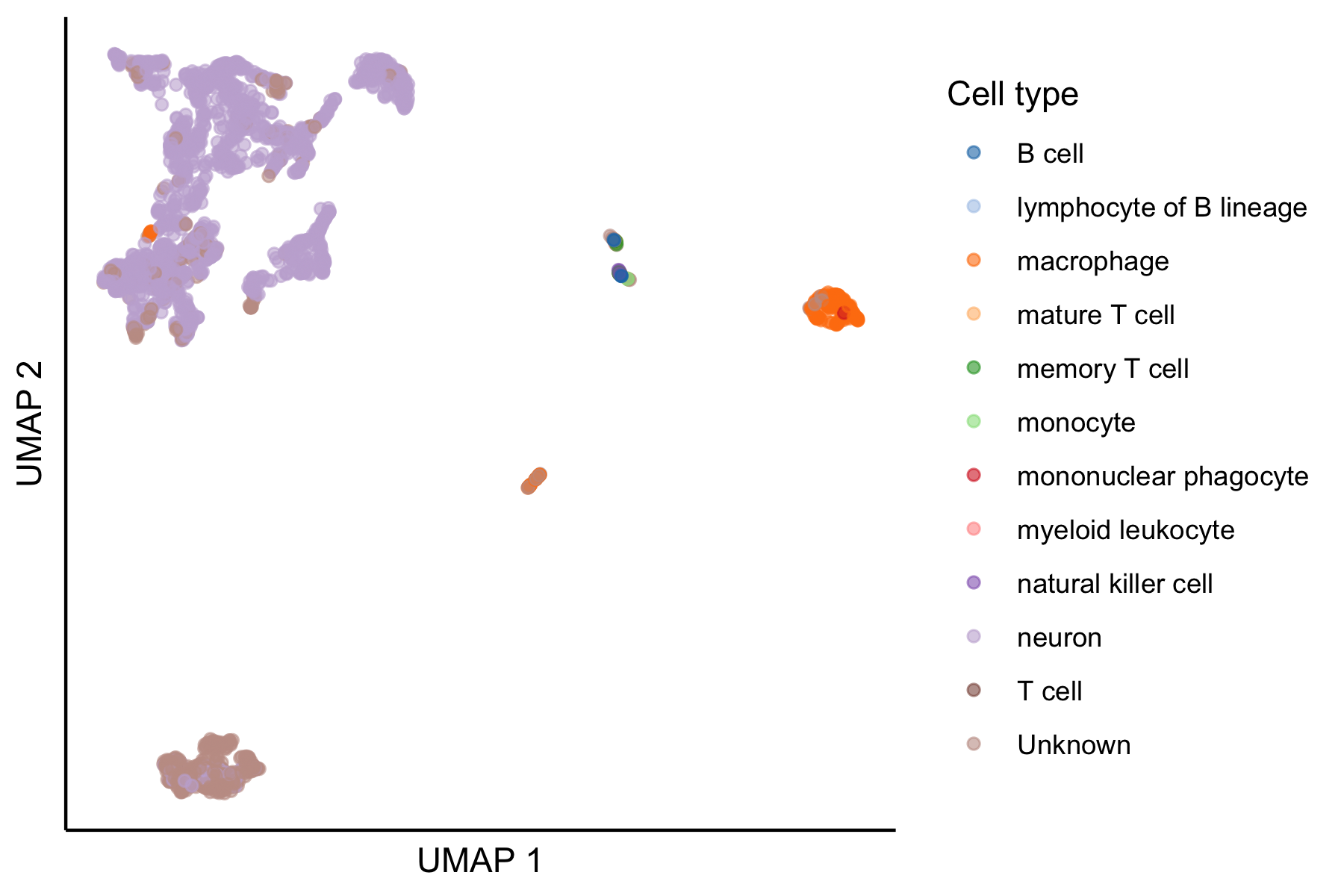
## ENSG00000186092 ENSG00000186092 OR4F5 0.000000000 0.0000000And of course we can make some standard plots, here a UMAP colored by the consensus cell type annotation.
# View the UMAP, colored by consensus cell type
scater::plotUMAP(sce, color_by = "consensus_celltype_annotation") +
ggplot2::guides(color = ggplot2::guide_legend(title="Cell type")) +
ggplot2::theme_classic() +
# remove axis ticks
ggplot2::theme(
axis.ticks = ggplot2::element_blank(),
axis.text = ggplot2::element_blank()
)
Downloading data in AnnData/H5AD format
ScPCAr also supports downloading data in AnnData (H5AD)
format, which is commonly used in Python-based single-cell analysis
workflows:
# Download the same sample in H5AD format
file_paths_h5ad <- ScPCAr::download_sample(
sample_id = sample_id,
auth_token = auth_token,
destination = "scpca_data",
format = "anndata"
)For more about the expected files when downloading samples in this format, see the ScPCA Portal AnnData docs.
Downloading projects
In addition to downloading the data for a single sample,
ScPCAr provides a function to download an entire project,
fittingly named download_project(). This function takes a
project id and authentication token as required input, and like the
download_sample function allows you to specify the
destination and format of the downloaded files. There are also a few
other options, such as the ability to download a merged object
containing all samples in the project, and whether to include
multiplexed samples, where multiple samples were pooled and sequenced
together, but have not been demultiplexed. Please see the function
documentation for more information.
# Download an entire project in SingleCellExperiment format,
# with separate files for each sample (default behavior).
project_file_paths <- ScPCAr::download_project(
project_id = project_id,
auth_token = auth_token,
destination = "scpca_data",
format = "sce"
)
# Download an entire project in SingleCellExperiment format,
# with all samples merged into a single object (but not integrated!).
project_file_paths <- ScPCAr::download_project(
project_id = project_id,
auth_token = auth_token,
destination = "scpca_data",
format = "sce",
merged = TRUE
)Session info
Click to expand R session info
## R version 4.4.3 (2025-02-28)
## Platform: aarch64-apple-darwin20
## Running under: macOS Sequoia 15.7.1
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## time zone: America/New_York
## tzcode source: internal
##
## attached base packages:
## [1] stats4 stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] SingleCellExperiment_1.28.1 SummarizedExperiment_1.36.0
## [3] Biobase_2.66.0 GenomicRanges_1.58.0
## [5] GenomeInfoDb_1.42.3 IRanges_2.40.1
## [7] S4Vectors_0.44.0 BiocGenerics_0.52.0
## [9] MatrixGenerics_1.18.1 matrixStats_1.4.1
## [11] ScPCAr_0.1.0 testthat_3.2.3
##
## loaded via a namespace (and not attached):
## [1] gridExtra_2.3 httr2_1.2.1 remotes_2.5.0
## [4] rlang_1.1.6 magrittr_2.0.3 scater_1.34.1
## [7] compiler_4.4.3 vctrs_0.6.5 stringr_1.5.1
## [10] profvis_0.4.0 pkgconfig_2.0.3 crayon_1.5.3
## [13] fastmap_1.2.0 XVector_0.46.0 ellipsis_0.3.2
## [16] labeling_0.4.3 scuttle_1.16.0 utf8_1.2.5
## [19] promises_1.3.0 sessioninfo_1.2.2 tzdb_0.5.0
## [22] UCSC.utils_1.2.0 ggbeeswarm_0.7.2 purrr_1.0.2
## [25] xfun_0.52 zlibbioc_1.52.0 cachem_1.1.0
## [28] beachmat_2.22.0 jsonlite_2.0.0 later_1.3.2
## [31] DelayedArray_0.32.0 BiocParallel_1.40.2 irlba_2.3.5.1
## [34] parallel_4.4.3 R6_2.6.1 stringi_1.8.7
## [37] pkgload_1.4.0 brio_1.1.5 Rcpp_1.0.13
## [40] knitr_1.50 usethis_3.1.0 readr_2.1.5
## [43] httpuv_1.6.15 Matrix_1.7-0 tidyselect_1.2.1
## [46] viridis_0.6.5 rstudioapi_0.17.1 abind_1.4-8
## [49] yaml_2.3.10 codetools_0.2-20 miniUI_0.1.1.1
## [52] curl_7.0.0 pkgbuild_1.4.6 lattice_0.22-6
## [55] tibble_3.2.1 shiny_1.9.1 withr_3.0.2
## [58] evaluate_1.0.3 desc_1.4.3 urlchecker_1.0.1
## [61] pillar_1.10.2 generics_0.1.4 rprojroot_2.0.4
## [64] hms_1.1.3 ggplot2_3.5.1 munsell_0.5.1
## [67] scales_1.3.0 xtable_1.8-4 glue_1.8.0
## [70] tools_4.4.3 BiocNeighbors_2.0.1 ScaledMatrix_1.14.0
## [73] fs_1.6.6 cowplot_1.1.3 grid_4.4.3
## [76] tidyr_1.3.1 devtools_2.4.5 colorspace_2.1-1
## [79] GenomeInfoDbData_1.2.13 beeswarm_0.4.0 BiocSingular_1.22.0
## [82] vipor_0.4.7 cli_3.6.5 rsvd_1.0.5
## [85] rappdirs_0.3.3 viridisLite_0.4.2 S4Arrays_1.6.0
## [88] dplyr_1.1.4 gtable_0.3.5 digest_0.6.37
## [91] ggrepel_0.9.6 SparseArray_1.6.2 farver_2.1.2
## [94] htmlwidgets_1.6.4 memoise_2.0.1 htmltools_0.5.8.1
## [97] lifecycle_1.0.4 httr_1.4.7 here_1.0.1
## [100] mime_0.13